Unwrapping the mystery of ancient Egyptian mummies
Reviving an ancient Egyptian Mummy sounds like something out of a science fiction movie, but researchers at the University of Melbourne have done the next best thing. In a multidisciplinary project with the Faculty of Medicine, Dentistry and Health Sciences headed by Dr Varsha Pilbrow, the head of Meritamun – a young Egyptian woman who lived more than 2000 years ago – has been imaged using CT and reconstructed using 3D-printing technology.
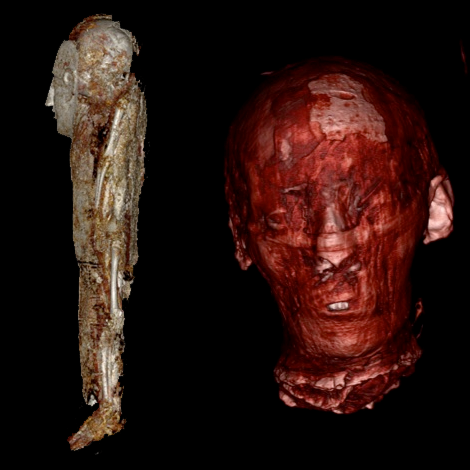
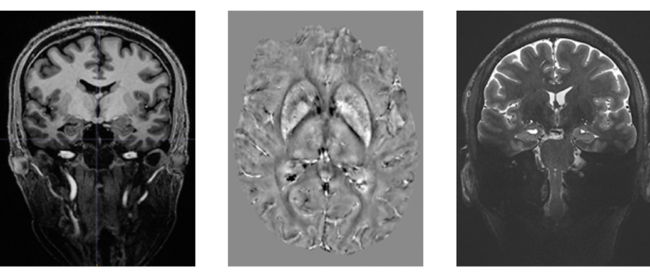
On the left, CT reconstructions of a baby mummy (still being studied). On the right, the CT reconstruction of the head of the mummy named Meritamun from the University of Melbourne’s collection within the School of Biomedical Sciences
Without disturbing the rare specimen and adhering to the controls and procedures of the Human Tissue Act 1982, Meritamun was imaged using the Siemens Human PET/CT in the University of Melbourne National Imaging Facility (NIF) Node. Digital and volumetric displays of tomographic data were acquired and reconstructed for 3D printing to create a skull replica.
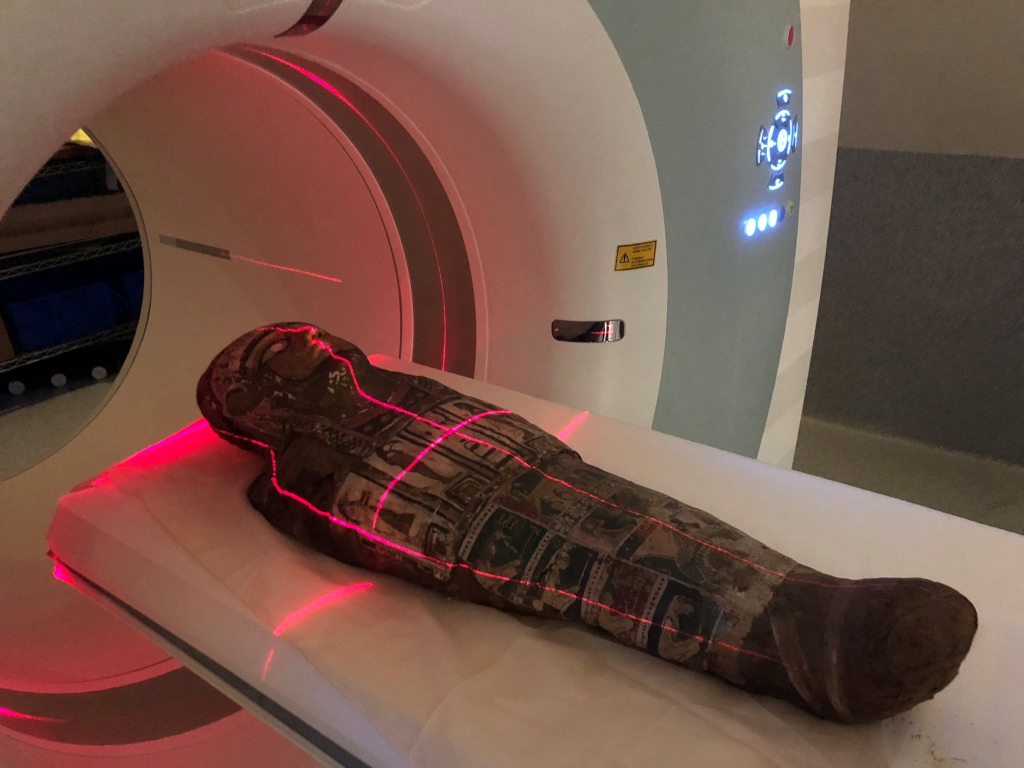
The scanning of a mummy with no adverse affects
Biomedical science Masters Student Stacey Gorski has used the CT data to diagnose Meritanum with anaemia, and hypothesizes a cause of death due to parasitic infection. “The fact that she lived to adulthood suggests that she was infected later in life,” says Gorski. She and supervisor Dr Pilbrow are continuing the investigation, hoping to learn more about the life and death of the ancient Egyptian using forensic pathology.
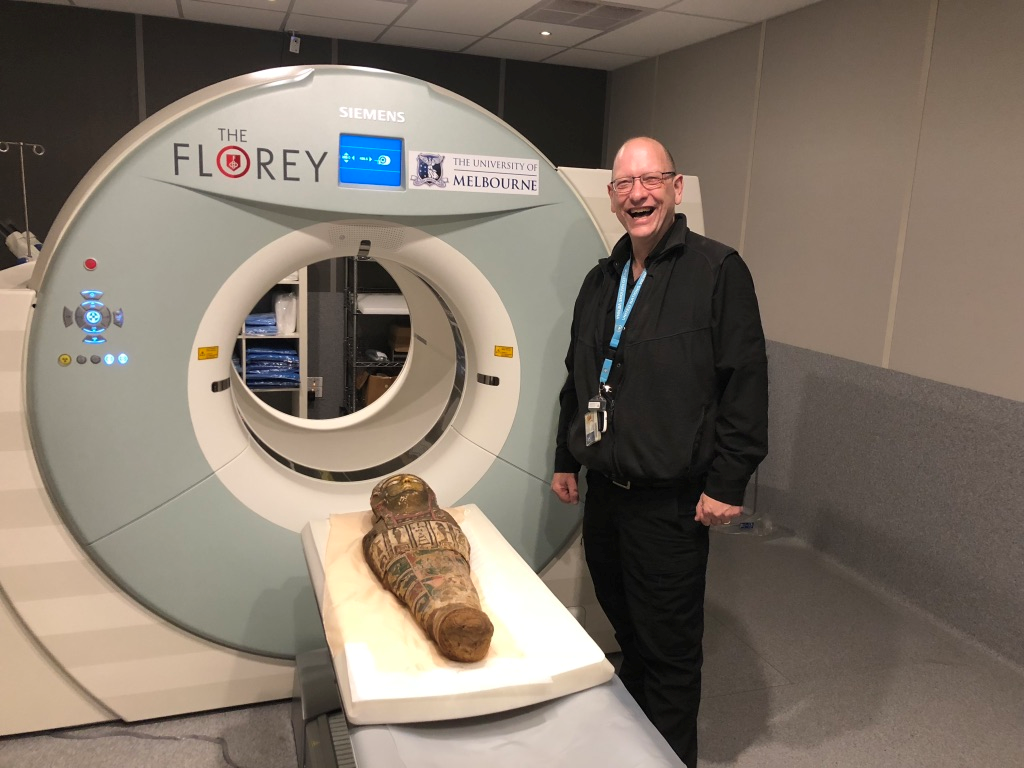
NIF Facility Fellow Mr Rob Williams facilitating access and providing expertise to the Human PET/CT scanner
In addition to learning more about the pathology and environments of population groups of 2000 years ago, the capability to replicate body parts and organs from CT imaging of specimens offers an opportunity for students to interact with old rare samples without damage to the original.
This story was contributed by the University of Melbourne. Acknowledgements go to Varsha Pilbrow, Julietta Capodistrias, Nina Sellars, Quentin Fogg, Michelle Gough, Gavan Mitchell, Peter Mayal, and Natalie Langowski.
For more information, please contact Rob Williams.