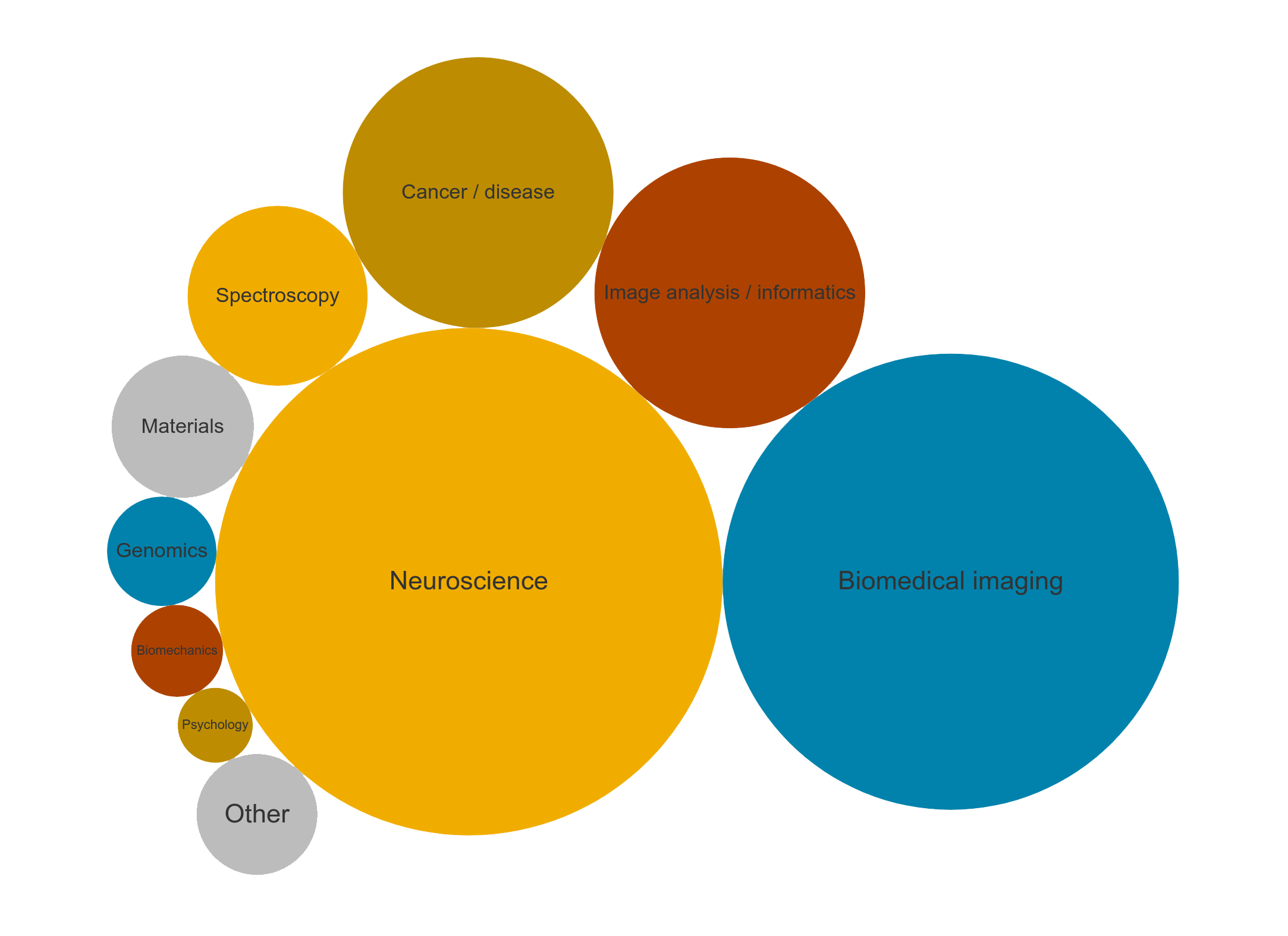
Magnetic Lymphatic Mapping in Pigs
Working Towards a New Gold Standard for Cancer Care
The lymphatic system is one of the key mechanisms for metastatic spread, whereby cancer cells that have disseminated from a primary tumour are taken up into lymphatic vessels and transported to other locations in the body, beginning with the lymph nodes. Conventionally, surgeons map the lymphatic flow from a tumour site into the first draining nodes – known as sentinel lymph nodes – using a Tc99m radiocolloid. Drainage pathways of tracers may be imaged before surgery with gamma cameras (lymphoscintigraphy), and individual nodes detected during surgery with handheld gamma probes. These sentinel nodes are the ‘canary in the coal mine’ and can offer valuable information to help stage the disease and determine the most appropriate treatment options, so it is ideal for removing just these nodes while leaving uninvolved nodes intact.
While this technique is successfully and routinely applied to breast cancer and melanoma, accuracy is limited in cancers where the nodes are in close proximity to the primary tumour or other nodes, such as cancers of the head and neck. Without an accurate method to identify sentinel nodes in oral cancer, extensive dissection of all nodes in the neck region is routinely performed to ensure any potential draining nodes are harvested, yet approximately 75 % of patients undergoing neck dissection are exposed to the complications and morbidity of this invasive procedure without clinical benefit.
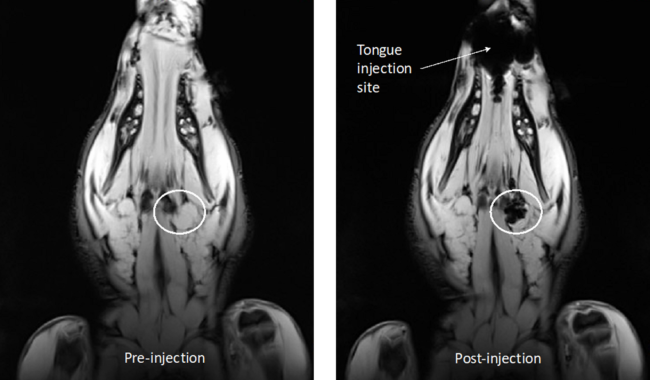
Dr Aidan Cousins and Dr Giri Krishnan have been working as part of a collaborative project between the University of South Australia and the University of Adelaide to demonstrate a novel, high-resolution technique for lymphatic mapping using magnetic nanoparticle-based tracers. Studies conducted at the South Australian Health and Medical Research Institute (SAHMRI) involve the injection of the magnetic tracer to the oral cavity of female Large White pigs, which are then scanned with a 3.0T Siemens MRI located at the large animal research and imaging facility (LARIF) NIF Node. Post-injection scans give Dr Cousins and Dr Krishnan detailed anatomical information of the drainage patterns of the tracer, which is then used to plan the surgery. During surgery, a handheld magnetometer probe developed by Dr Cousins is used, along with MRI data and visual identification, to pinpoint the draining nodes of interest from other, uninvolved nodes.
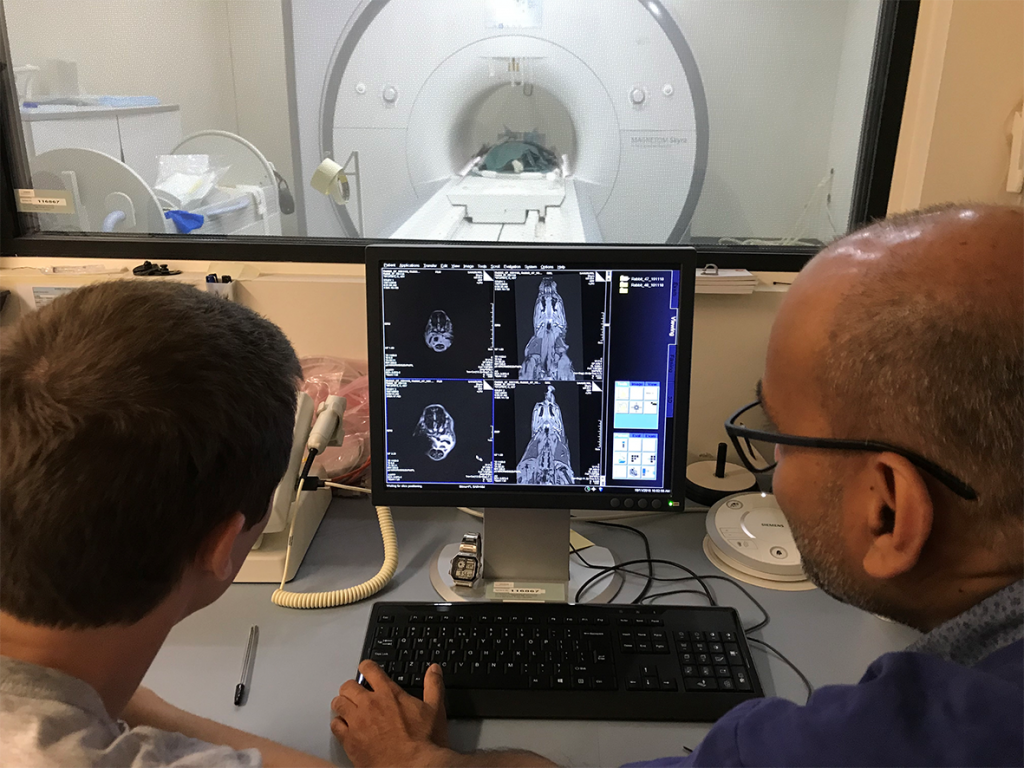
Dr Cousins with Raj Perumal (LARIF) examining an MRI scan of a pig following injection of the magnetic tracer
The results of this experiment showed the magnetic alternative to be adept for mapping lymphatic drainage in complex environments. Triangulating the location draining nodes with high precision before surgery was made possible by the high-resolution soft-tissue detail afforded by the 3.0T MRI. During surgery, the handheld probe was able to identify all draining nodes by way of detecting their magnetic ‘signature’. This final confirmation of draining nodes is analogous to the use of handheld gamma probes but has the distinct advantage of pin-point resolution, meaning nodes can be in very close (touching) proximity to each other and it is still possible to differentiate between the individual nodes’ magnetic signals. This spatial resolution is currently unmatched by any other probe technology commercially available and is key to the application of sentinel node mapping in complex cancers.

Photo of the high-resolution magnetometer probe developed by Dr Cousins
Results from this study are being used to design a world-first clinical trial applying magnetic tracers and a high-resolution probe to human patients with cancer of the oral cavity.
This story was contributed by SAHMRI. For further inquiries, please contact Mr Raj Perumal