Closing the acquisition time gap – a new sequence scheme for optimised Sodium MRI
Magnetic Resonance (MR) opens a window for the non-invasive investigation of MR-observable nuclei, i.e. nuclei with a non-zero spin. Common nuclei in organic substances, like Carbon-12 and Oxygen-16; however, possess a zero-spin consequently rendering Hydrogen-1 (commonly referred as ‘proton‘ due to the single proton in the nucleus) the first and most frequently imaged nuclei.
Despite the much lower signal regime, also the potential of imaging ‘other nuclei’, so-called x-Nuclei MRI was approached in the early years of MRI. In 1985, only a few years after the first proton-based images were published, Sodium-23 was the second nucleus in the human body that was imaged non-invasively using MRI by Hilal and coworkers [1]. In spite of these early beginnings, x-Nuclei imaging in general and Sodium MRI, in particular, did not see the same high-paced progression as proton-based MRI techniques. This downturn was mostly caused by significant SNR limitations resulting from the low in vivo concentration and complicated NMR-signal characteristics of Sodium nuclei.
While SNR issues are more and more overcome through hardware improvements, particularly the recent advent of high field systems, the challenging signal characteristics continue to encourage the development of dedicated acquisition methods. Sodium possesses a 3/2-spin and as such exhibits a fast bi-exponential signal decay. Consequently, Sodium MRI sequences are commonly performed in 3D with k-space sampled via centre-out trajectory schemes resulting in inherent drawbacks for sampling efficiency and SNR. While previous research in Sodium MRI method development focused on the optimisation of sampling trajectories, we present a new acquisition method that tackles Sodium image quality improvements via a sequence timing optimisation.
Recently, we introduced zero-gradient excitation ramped hybrid encoding (zGRF-RHE) Sodium MRI, a timing optimisation that utilises the sequence dead-time delay (time-gap between transmit and receive mode) for gradient pre-ramping. This concept improves encoding time across k-space and in so doing alleviates signal decay effects during the acquisition. It utilises a hybrid sequence encoding approach where the central k-space gap, resulting from gradient activity before receiver onset, is filled with Cartesian single point acquisitions. A sequence diagram of zGRF-RHE is displayed in Figure 1.
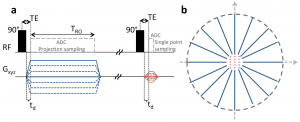
Figure 1: Radial zero‐gradient‐excitation ramped hybrid encoding (zGRF‐RHE) sequence diagram (A) and 2D illustration of hybrid k‐space encoding scheme (B). Gradient‐free excitation is followed by an immediate gradient ramping, data sampling commences after dead‐time. Central k-space is measured separately as single points with the same gradient pre‐ramping condition. TRO, readout duration; td, dead‐time. Figure taken from [2].
Hybrid imaging techniques, like PETRA or its generalised concept RHE, are frequently used in short-T2 proton imaging. Their application to low SNR problems like Sodium was, however, hampered by the employed gradient modulation during signal excitation. In that regard, we establish zero-gradient excitation for an artefact-free excitation profile maximizing image quality. The gradient-free excitation supports high flip angle acquisition, an essential requirement for low SNR imaging like Sodium, but also provides the opportunity to employ advanced RF pulse shapes without introducing a gradient-based excitation bias. Additionally, it should be highlighted that the zGRF-RHE sequence approach essentially describes a timing concept that is not just independent of the RF-excitation but also is not restricted to a particular readout-scheme.
An investigation of 3D-radial zGRF-RHE with standard 3D-radial ultra-short echo time (UTE) imaging can be found in our recent publication [2]. Compared to simulations, phantoms and in-vivo human brain acquisitions confirmed that our proposed sequence timing concept improves on image quality independent of the readout bandwidth and shows reduced image blurring and higher SNR. Figure 2 displays results from an in vivo human brain experiment highlighting improved image sharpness.
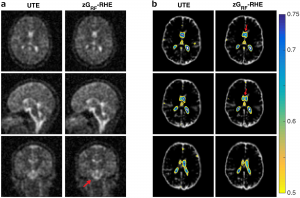
Figure 2: (A) Cross sections through matched slices in in vivo experiments at TRO of 2 ms UTE and zGRF‐RHE. zGRF‐RHE shows sharper details around the brain stem (red arrow). (B) Three consecutive transverse slices of CSF masks with contour lines of normalized UTE and zGRF‐RHE at TRO of 2 ms. zGRF‐RHE provides better delineation between lateral ventricles (red arrow). Figure taken from [2].
In summary, this work introduces an enhanced sequence timing concept with particular applicability to challenging low SNR-problems. Ultimately, this approach is expected to be of use for a wider range of x-Nuclei applications with spin > ½.
For further information, the interested reader is pointed to our full publication which can be accessed at https://onlinelibrary.wiley.com/doi/abs/10.1002/mrm.27484.
References
[1] Hilal, S. K., Maudsley, A. A., Ra, J., Simon, H. E., Roschmann, P., Wittekoek, S., Cho, Z., and Mun, S. (1985). In vivo NMR imaging of sodium-23 in the human head. Journal of Computer Assisted Tomography, 9(1):1–7.
[2] Blunck Y, Moffat BA, Kolbe SC, Ordidge RJ, Cleary JO, Johnston LA. Zero-Gradient-Excitation Ramped Hybrid Encoding (zGRF-RHE) Sodium MRI. Magnetic Resonance in Medicine 2018. https://doi.org/10.1002/mrm.27484.
This story was contributed by Yasmin Blunck of the Melbourne Brain Centre Imaging Unit and Department of Biomedical Engineering, University of Melbourne, Parkville


