MRtrix3
MRtrix3: Advanced tools for the analysis of diffusion MRI data
Diffusion-weighted MRI (dMRI) is a commonly-used medical imaging modality for the investigation of tissue microstructure, exploiting the local hindrance and restriction of water diffusion as indirect probes. The neuroimaging research community utilises this technology extensively for the study of brain white matter in particular, reconstructing structural connectivity pathways and analysing estimated tissue properties.
To do so requires extensive complex computations, including image pre-processing, biophysical modelling, digital reconstruction of neuronal fibre trajectories, and tailored statistical analysis and visualisation tools. The quality and capabilities of neuroscience research utilising this technology – as well as other applications such as neurosurgical planning – rely on all utilised aspects of this pipeline being robust yet powerful.
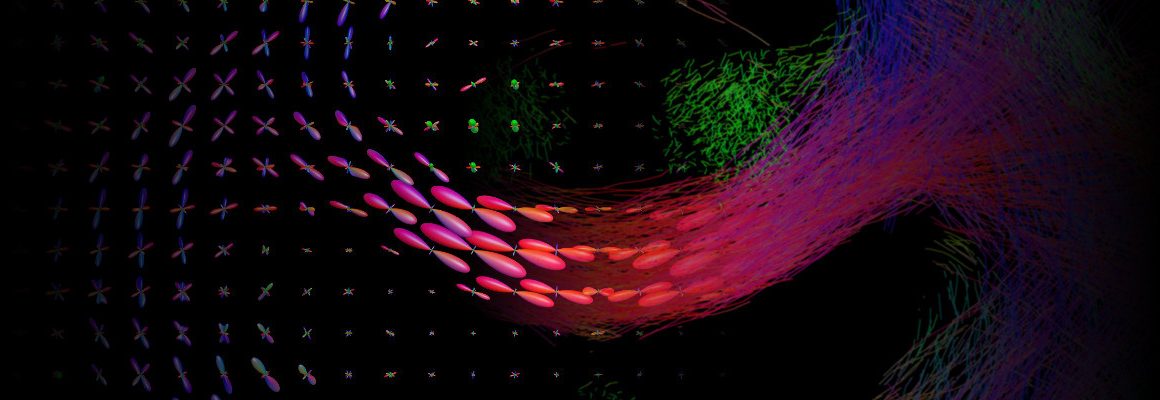
The recently released MRtrix 3.0.0 is a free, open-source software package that incorporates various state-of-the-art technologies in the field of dMRI, combining high computational performance with a consistent, user-friendly interface. It is built principally around the “spherical deconvolution” model, which is regularly utilised in the field as the reference model for optimal resolution of the various complex fibre configurations found throughout the brain white matter. Additional tools are provided for both quantitative estimations of structural connectivity between brain grey matter regions, and data-driven hypothesis testing of quantitative white matter imaging metrics, in the presence of such tissue complexity.
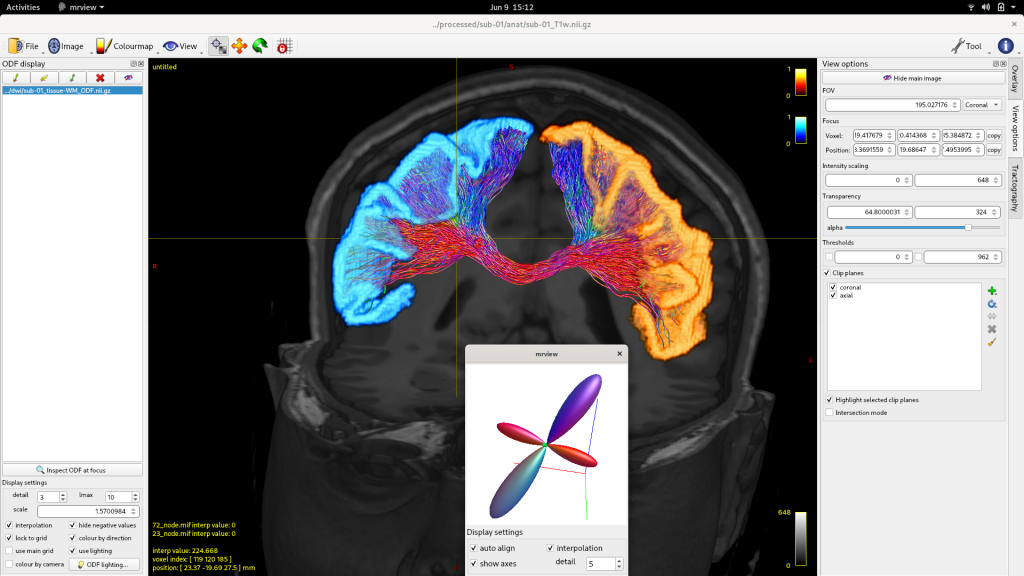
An international collaboration of researchers has recently published the implementation of MRtrix3 in the peer-reviewed journal, NeuroImage. Lead author Dr J-Donald Tournier and colleagues provide “a high-level overview of the features of the MRtrix3 framework and general-purpose image processing applications provided with the software”, highlighting the guiding principles of development including reproducible neuroscience and consistent documentation. This manuscript also details the underlying software framework and Application Programming Interface (API) of the MRtrix3 software to encourage utilisation of its library features by other neuroscience methods developers.
NIF Facility Fellow Dr Robert Smith, of the Florey Node, is one of two principal developers of this software package, with over 3,200 code commits and 220,000 lines of code contributed. His involvement with the software project began not long after the commencement of his PhD candidature in 2009, for which Dr Tournier was a supervisor. The two key technical developments resulting from his PhD – the “Anatomically-Constrained Tractography (ACT)” and “Spherical-deconvolution Informed Filtering of Tractograms (SIFT)” methods (each of which has over 250 citations on Google Scholar) were developed & distributed with the MRtrix3 package, improving the biological accuracy and quantitative properties of white matter connectivity experiments throughout the worldwide neuroscience community.

The MRtrix3 community forum, where users of the software interact directly with developers and one another, currently has over 900 active users and over 1.5 million page views. The manuscript describing the previous version of this software (“MRtrix 0.2”), published in 2012, now has over 500 citations; while the MRtrix3 manuscript was published too recently to accumulate a large citation count, its long-term influence would be reasonably expected to exceed this. Google Scholar searches yield over 1500 hits for “mrtrix diffusion mri”, and over 450 hits for “mrtrix surgery”. Some of the capabilities included in MRtrix3 are incorporated in Siemens’ Track Density Imaging and tractography prototype, currently available for early testing at participating Siemens MRI research sites. The software is also utilised by many companies that specialise in providing third-party services for image processing and analysis. The development team additionally runs software workshops to teach usage of the software to the research community.
The tools provided within this software package can improve the accuracy and utility of diffusion MRI analysis in many contexts, both neuroscientific and clinical. This will, over time, include characterisation of various brain disorders, tracking the efficacy of potential treatments of such, neurosurgical planning, and diagnosis.
This story was contributed by Dr Robert Smith. For more information, please visit the MRtrix3 website, community forum, GitHub repository, or follow on Twitter.